SDS-WAS regional dust forecasts#
This notebook provides an introduction to dust forecast data from the MONARCH model. The notebook introduces you to the variable Dust Optical Depth and you will learn how the model has predicted the Saharan Dust event which occured over Europe in the first half of April 2024.
The WMO Sand and Dust Storm Warning Advisory and Assessment System (SDS-WAS). is an international framework linking institutions involved in Sand and Dust Storm (SDS) research, operations and delivery of services.
The framework is organised in several regional centers, which aim to implement SDS-WAS objectives in a specific region. The Barcelona Supercomputing Center (BSC-CNS) and the Meteorological State Agency of Spain (AEMET) are hosting the SDS-WAS regional center for Northern Africa, Middle East and Europe.
One of the main activities of the SDS-WAS regional center is to provide daily operational dust forecasts for Northern Africa (north of the equator), Middle East and Europe. The BCS-CNS, in collaboration with NOAA’s National Centers for Environmental Prediction (NCEP), the NASA’s Goddard Institute for Space Studies and the International Research Institute for Climate Society (IRI), has developed MONARCH, an online multi-scale atmospheric dust model intended to provide short and medium-range dust forecasts for both, regional and global domains.
The model provides forecast information up to 72 hours in advance (every 3 hours) of two parameters: Dust Optical Depth and Dust Surface Concentration.
MONARCH Forecast data are available in netCDF format and are available for download via a THREDDS Data Server here.
Basic facts
Spatial resolution: 0.1° x 0.1°
Spatial coverage: Northern Africa, Middle East and Europe
Temporal resolution: 3-hourly up to 72 hours in advance
Temporal coverage: since February 2012
Data format: NetCDF
How to access the data
Dust forecast data from the MONARCH model are available for download via the website of the WMO Barcelona Dust Regional Center.
In order to be able to download data from the portal, you need to register via this contact form. Below, you see an example how you can programmatically download one data file.
Learn more about the data products and how to access them from this user guide.
Load required libraries
import xarray as xr
import requests
import matplotlib.pyplot as plt
import matplotlib.colors
from matplotlib.cm import get_cmap
from matplotlib import animation
from matplotlib.axes import Axes
import cartopy.crs as ccrs
from cartopy.mpl.gridliner import LONGITUDE_FORMATTER, LATITUDE_FORMATTER
import cartopy.feature as cfeature
from cartopy.mpl.geoaxes import GeoAxes
GeoAxes._pcolormesh_patched = Axes.pcolormesh
from IPython.display import HTML
import warnings
warnings.simplefilter(action = "ignore", category = RuntimeWarning)
Load helper functions
%run ../functions.ipynb
Download MONARCH dust forecast data#
Data products from the WMO Barcelona Dust Regional Center can be accessed via a THREDDS Data Server. After registration, you received access credentials that you can use to access the data files. You can store your access credentials as variables, user and password respectively. Note: you have to replace the ##### with your credentials.
user='################'
password='###################'
The data catalog offers public and restricted data. MONARCH dust forecasts are available as public data files. To get the download link for the dust forecast issued on 7 April 2024, you have to navigate to the following folder in the data catalog: /BDRC_THREDDS_Public_Data/MONARCH/2024/04/. If you click on the file for 7 April 2024, you get different download options. In the following, you see an example how you can use the Python library requests to download data from a given url.
First, let us store the url path of the data file as variable.
url='https://dust.aemet.es/thredds/fileServer/dataRoot/MONARCH/2024/04/2024040712_3H_SDSWAS_NMMB-BSC-v2_OPER.nc'
Next, you can now establish a connection to the server where the data file is hosted, with the function get() from the requests package. You have to specify auth as keyword argument and give the function your access credentials (user, password).
Once a successful connection is established, you can open a new file and write the content of the server connection. The result is a netCDF data with a size of ~900 MB.
r = requests.get(url, auth=(user,password))
open('./20240407_MONARCH_dust_forecast.nc', 'wb').write(r.content)
Load and browse MONARCH dust forecast data#
The first step is to load a MONARCH forecast file to better understand its structure. The data is disseminated in the netCDF format on a daily basis, with the forecast initialisation at 12:00 UTC. Let us load the MONARCH dust forecast of 7 April 2024. You can use the function open_dataset() from the xarray Python library, which makes reading a netCDF file very efficient and easy. The function loads the netCDF file as xarray.Dataset(), which is a data collection of multiple variables sharing the same coordinate information.
Below, you see that the MONARCH data has three dimensions, lat, lon and time, and offers two data variables od550_dust and sconc_dust.
filepath = '../eodata/3_model/sds_was/20240407_MONARCH_dust_forecast.nc'
file = xr.open_dataset(filepath)
file
<xarray.Dataset> Size: 947MB
Dimensions: (time: 29, lat: 825, lon: 1650)
Coordinates:
* lat (lat) float64 7kB -10.95 -10.85 -10.75 ... 71.25 71.35 71.45
* lon (lon) float64 13kB -62.95 -62.85 -62.75 ... 101.8 101.9 102.0
* time (time) datetime64[ns] 232B 2024-04-07T12:00:00 ... 2024-04-11
Data variables:
dust_ext_sfc (time, lat, lon) float32 158MB ...
dust_load (time, lat, lon) float32 158MB ...
dust_depd (time, lat, lon) float32 158MB ...
dust_depw (time, lat, lon) float32 158MB ...
sconc_dust (time, lat, lon) float32 158MB ...
od550_dust (time, lat, lon) float32 158MB ...
Attributes:
Domain: Regional
Conventions: CF-1.7
comment: Generated on cirrus
NCO: netCDF Operators version 4.8.1 (Homepage = ht...
history: Mon Apr 8 23:00:59 2024: ncap2 -O -v -s wher...
history_of_appended_files: Sun Apr 7 18:18:23 2024: Appended file od550...Let us now have a closer look at the dimensions of the data. Let us first inspect the two coordinate dimensions lat and lon. You can simply access the dimension’s xarray.DataArray by specifying the name of the dimension. Below you see that the data has a 0.1 x 0.1 degrees resolution and have the following geographical bounds:
Longitude: [-63., 101.9]Latitude: [-11., 71.4]
latitude = file.lat
longitude = file.lon
latitude, longitude
(<xarray.DataArray 'lat' (lat: 825)> Size: 7kB
array([-10.95, -10.85, -10.75, ..., 71.25, 71.35, 71.45])
Coordinates:
* lat (lat) float64 7kB -10.95 -10.85 -10.75 -10.65 ... 71.25 71.35 71.45
Attributes:
units: degrees_north
axis: Y
long_name: latitude coordinate
standard_name: latitude,
<xarray.DataArray 'lon' (lon: 1650)> Size: 13kB
array([-62.95, -62.85, -62.75, ..., 101.75, 101.85, 101.95])
Coordinates:
* lon (lon) float64 13kB -62.95 -62.85 -62.75 ... 101.8 101.9 102.0
Attributes:
units: degrees_east
axis: X
long_name: longitude coordinate
standard_name: longitude)
Now, let us also inspect the time dimension. You see that one daily forecast file has 25 time steps, with three hourly forecast information up to 72 hours (3 days) in advance.
file.time
<xarray.DataArray 'time' (time: 29)> Size: 232B
array(['2024-04-07T12:00:00.000000000', '2024-04-07T15:00:00.000000000',
'2024-04-07T18:00:00.000000000', '2024-04-07T21:00:00.000000000',
'2024-04-08T00:00:00.000000000', '2024-04-08T03:00:00.000000000',
'2024-04-08T06:00:00.000000000', '2024-04-08T09:00:00.000000000',
'2024-04-08T12:00:00.000000000', '2024-04-08T15:00:00.000000000',
'2024-04-08T18:00:00.000000000', '2024-04-08T21:00:00.000000000',
'2024-04-09T00:00:00.000000000', '2024-04-09T03:00:00.000000000',
'2024-04-09T06:00:00.000000000', '2024-04-09T09:00:00.000000000',
'2024-04-09T12:00:00.000000000', '2024-04-09T15:00:00.000000000',
'2024-04-09T18:00:00.000000000', '2024-04-09T21:00:00.000000000',
'2024-04-10T00:00:00.000000000', '2024-04-10T03:00:00.000000000',
'2024-04-10T06:00:00.000000000', '2024-04-10T09:00:00.000000000',
'2024-04-10T12:00:00.000000000', '2024-04-10T15:00:00.000000000',
'2024-04-10T18:00:00.000000000', '2024-04-10T21:00:00.000000000',
'2024-04-11T00:00:00.000000000'], dtype='datetime64[ns]')
Coordinates:
* time (time) datetime64[ns] 232B 2024-04-07T12:00:00 ... 2024-04-11
Attributes:
standard_name: time
long_name: timeLoad data variables as data arrays#
A xarray.Dataset is a collection of multiple variables and offers a general overview of the data, but does not offer direct access to the data arrays. For this reason, you want to load a data variable as xarray.DataArray. You can access the data array information by simply specifying the name of the variable after the name of the xarray.Dataset
Let us load both variables as xarray.DataArray, od550_dust and sconc_dust. Below you see that, once the data array is loaded, that each variable is disseminated with additional attributes, e.g. long_name and units. These attributes give us more information about the data. od550_dust e.g. is Aerosol Optical Depth which does not have a unit and sconc_dust is the surface dust concentration measured in kg / m3.
od_dust = file.od550_dust
od_dust
<xarray.DataArray 'od550_dust' (time: 29, lat: 825, lon: 1650)> Size: 158MB
[39476250 values with dtype=float32]
Coordinates:
* lat (lat) float64 7kB -10.95 -10.85 -10.75 -10.65 ... 71.25 71.35 71.45
* lon (lon) float64 13kB -62.95 -62.85 -62.75 ... 101.8 101.9 102.0
* time (time) datetime64[ns] 232B 2024-04-07T12:00:00 ... 2024-04-11
Attributes:
grid_mapping: crs
units: -sconc_dust = file.sconc_dust
sconc_dust
<xarray.DataArray 'sconc_dust' (time: 29, lat: 825, lon: 1650)> Size: 158MB
[39476250 values with dtype=float32]
Coordinates:
* lat (lat) float64 7kB -10.95 -10.85 -10.75 -10.65 ... 71.25 71.35 71.45
* lon (lon) float64 13kB -62.95 -62.85 -62.75 ... 101.8 101.9 102.0
* time (time) datetime64[ns] 232B 2024-04-07T12:00:00 ... 2024-04-11
Attributes:
grid_mapping: crs
units: kgm-3You can define variables for the attributes of a variable. This can be helpful during data visualization, as e.g. the attribute units can be added as additional information to the plot. From the xarray.DataArray, you simply specify the name of the attribute of interest.
units= od_dust.units
Visualize dust AOD at 550 nm#
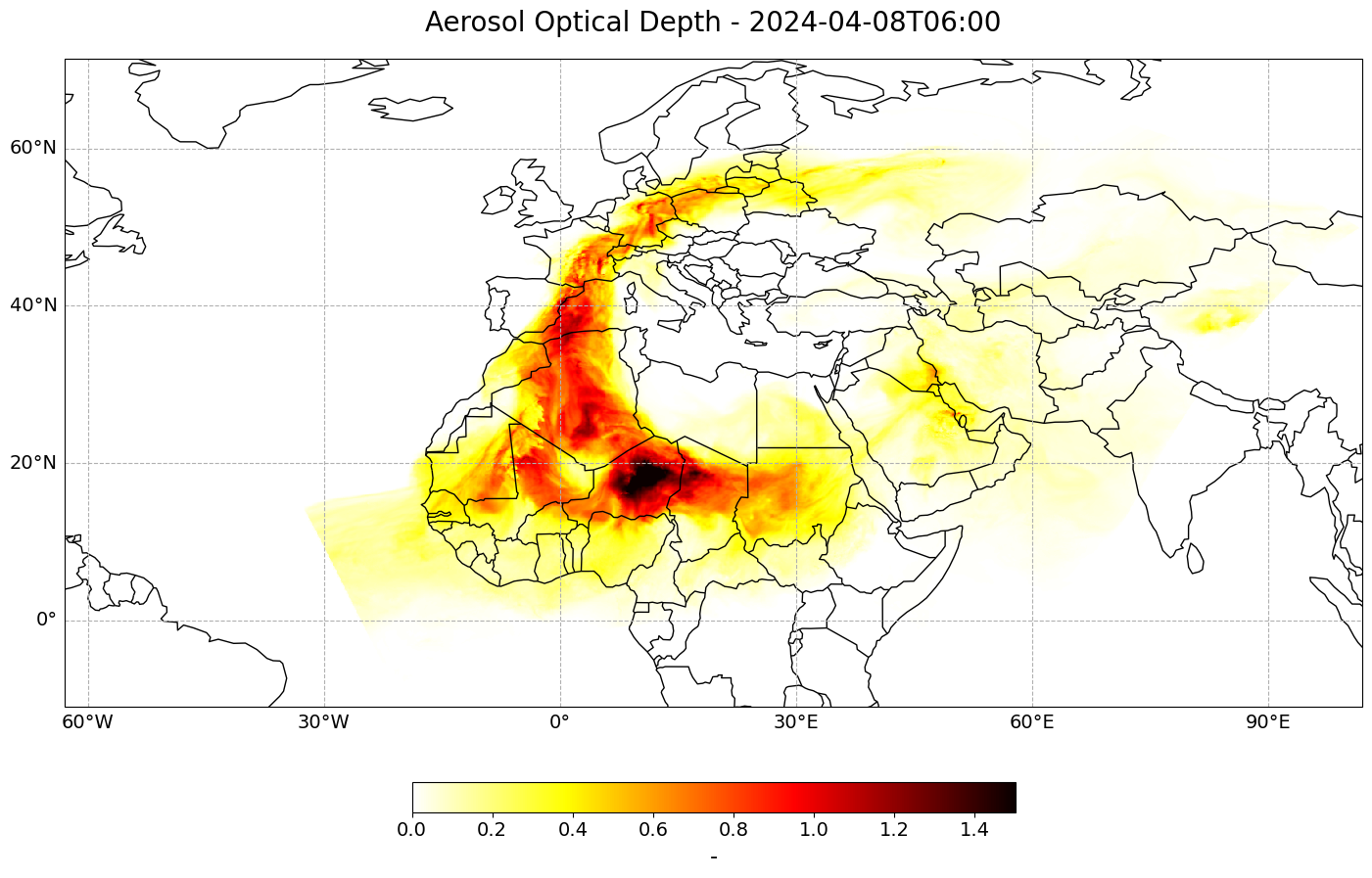
Now we have loaded all necessary information to be able to visualize the dust Aerosol Optical Depth for one specific time during the forecast run. Let us use the function visualize_pcolormesh() to visualize the data with the help of the plotting library matplotlib and Cartopy.
You have to specify the following keyword arguments:
data_array: thelongitude,latitude: longitude and latitude variables of the data variableprojection: one of Cartopy’s projection optionscolor_scale: one of matplotlib’s colormapsunits: unit of the data parameter, preferably taken from the data array’s attributeslong_name: longname of the data parameter is taken as title of the plot, preferably taken from the data array’s attributesvmin,vmax: minimum and maximum bounds of the color rangeset_global: False, if data is not globallonmin,lonmax,latmin,latmax: kwargs have to be specified, ifset_global=False
Note: in order to have the time information as part of the title, we add the string of the datetime information to the long_name variable: long_name + ' ' + str(od_dust[##,:,:].time.data)[0:16].
forecast_step = 6
visualize_pcolormesh(data_array=od_dust[forecast_step,:,:],
longitude=od_dust.lon,
latitude=od_dust.lat,
projection=ccrs.PlateCarree(),
color_scale='hot_r',
unit=units,
long_name='Aerosol Optical Depth - ' + str(od_dust[forecast_step,:,:].time.data)[0:16],
vmin=0,
vmax=1.5,
set_global=False,
lonmin=longitude.min().data,
lonmax=longitude.max().data,
latmin=latitude.min().data,
latmax=latitude.max().data)
(<Figure size 2000x1000 with 2 Axes>,
<GeoAxes: title={'center': 'Aerosol Optical Depth - 2024-04-08T06:00'}>)
Animate dust AOD forecasts#
In the last step, you can animate the Dust Aerosol Optical Depth forecasts in order to see how the trace gas develops over a period of 3 days, from 7th April 2024 12 UTC to 10th April 2024.
You can do animations with matplotlib’s function animation. Jupyter’s function HTML can then be used to display HTML and video content.
The animation function consists of 4 parts:
Setting the initial state:
Here, you define the general plot your animation shall use to initialise the animation. You can also define the number of frames (time steps) your animation shall have.Functions to animate:
An animation consists of three functions:draw(),init()andanimate().draw()is the function where individual frames are passed on and the figure is returned as image. In this example, the function redraws the plot for each time step.init()returns the figure you defined for the initial state.animate()returns thedraw()function and animates the function over the given number of frames (time steps).Create a
animate.FuncAnimationobject:
The functions defined before are now combined to build ananimate.FuncAnimationobject.Play the animation as video:
As a final step, you can integrate the animation into the notebook with theHTMLclass. You take the generate animation object and convert it to a HTML5 video with theto_html5_videofunction
# Setting the initial state:
# 1. Define figure for initial plot
fig, ax = visualize_pcolormesh(data_array=od_dust[0,:,:],
longitude=od_dust.lon,
latitude=od_dust.lat,
projection=ccrs.PlateCarree(),
color_scale='hot_r',
unit=units,
long_name='Aerosol Optical Depth - ' + str(od_dust[0,:,:].time.data)[0:16],
vmin=0,
vmax=1.5,
lonmin=longitude.min().data,
lonmax=longitude.max().data,
latmin=latitude.min().data,
latmax=latitude.max().data,
set_global=False)
frames = 24
def draw(i):
img = plt.pcolormesh(od_dust.lon,
od_dust.lat,
od_dust[i,:,:],
cmap='hot_r',
transform=ccrs.PlateCarree(),
vmin=0,
vmax=1.5,
shading='auto')
ax.set_title('Aerosol Optical Depth - '+ str(od_dust.time[i].data)[0:16], fontsize=20, pad=20.0)
return img
def init():
return fig
def animate(i):
return draw(i)
ani = animation.FuncAnimation(fig, animate, frames, interval=800, blit=False,
init_func=init, repeat=True)
HTML(ani.to_html5_video())
plt.close(fig)
HTML(ani.to_html5_video())

